Machine learning methods were applied on sequenced fungal genomes to classify gene families.
The Science
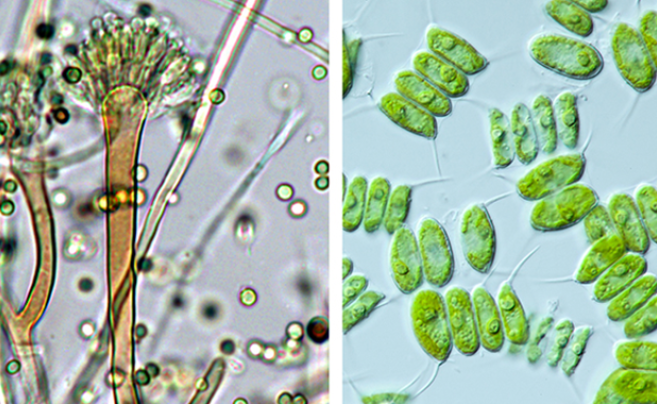
As a proof of concept, researchers developed an algorithm that “taught” a computer how to classify 101 representative genomes of Dothideomycetes, the largest class of fungi, by lifestyles. The machine “learned” to identify data generated in part through the 1000 Fungal Genomes Project, including 55 newly-sequenced species.
The Impact
The class Dothideomycetes includes fungi that obtain nutrients from decaying organic matter (saprobes), and many plant pathogens known to infect most major food crops and feedstocks for biomass and biofuel production. Learning about the ecology and evolution of Dothideomycetes could provide more insights into how these fungi have adapted to stress and host specificity, particularly regarding the effects of climate change.
Summary
A team led by researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), has generated a more accurate phylogenetic tree tracking the evolution of Dothideomycetes fungi. The work appeared in the June issue of Studies in Mycology.
This work was enabled in part through JGI’s Community Science Program (CSP), as several approved proposals have contributed toward filling gaps on the fungal Tree of Life, including the 1000 Fungal Genomes Project, which aims to provide a reference genome for each of the fungal families. Researchers had access to over 100 Dothideomycetes genomes, just enough of a sample size to test if a computer algorithm could distinguish between fungal lifestyles of saprobes and pathogens based on the data provided. In this proof of concept test, the team reported that the algorithm was over 95% successful at classifying the fungal genomes
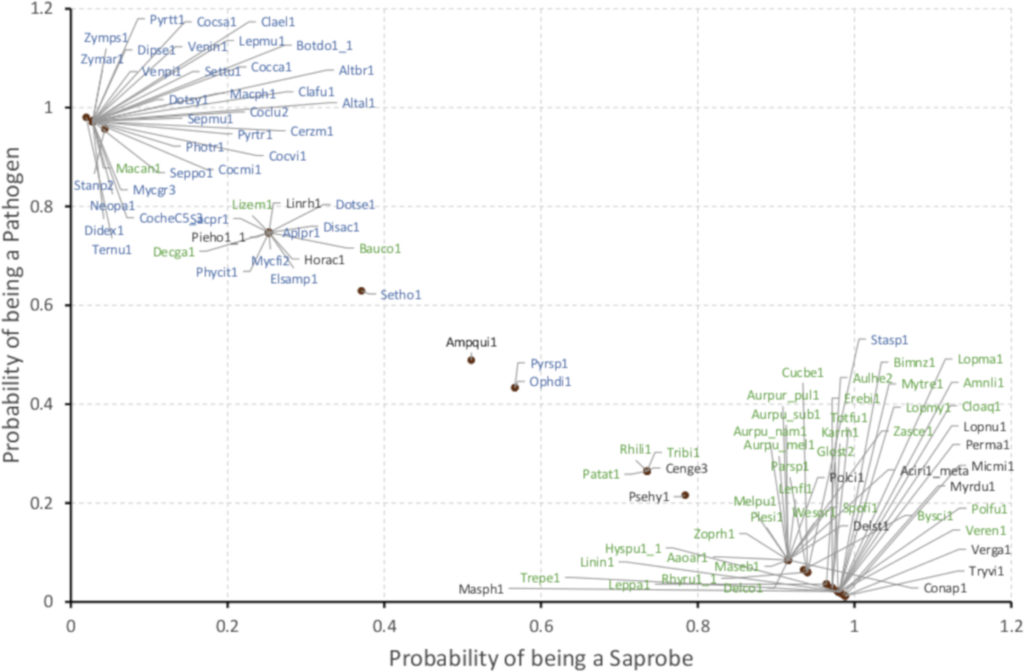
Support Vector Machine (SVM)-based prediction of lifestyle based on 6 gene clusters showed a >95 % accuracy in correctly predicting plant pathogens (blue) vs saprobes (green). (From Haridas S et al. Studies in Mycology, 2020.)
Using whole genome data from newly sequenced genomes, they also built a high-quality phylogenetic tree for Dothideomycetes. The data allowed the team to reclassify 25 Dothideomycetes species, and also provides more information about speciation events.