JGI researchers lend expertise to develop tools and data hub for NIH program.

While the microbes in, on and around Earth outnumber the stars in the Milky Way, viruses outnumber the microbes by at least another order of magnitude. For years, scientists at the DOE Joint Genome Institute, a DOE Office of Science user facility located at Lawrence Berkeley National Laboratory, have collaborated with global researchers on projects related to understanding how the interactions between viruses and their host microbes impact the regulation of global nutrient cycles. The JGI’s latest strategic plan (download a copy here) describes how understanding these nutrient cycles can help researchers contribute toward developing the bioeconomy.
Now, JGI researchers are lending their expertise in developing and managing public viral genomics resources such as analytical tools and data repositories to two projects funded through the National Institutes of Health’s (NIH) Common Fund Human Virome Program. Nikos Kyrpides, head of the JGI’s Microbiome Data Science, leads one project with JGI colleagues Antonio Camargo and Simon Roux. Kyrpides is part of another project led by Owen White at the University of Maryland, Baltimore. Both projects will build on the researchers’ expertise in developing and maintaining the JGI’s Integrated Microbial Genomics and Microbiomes (IMG/M) system.
The Program aims to better understand the viruses that coexist with humans – the “healthy” human virome - not just those associated with disease, similar to how the Human Microbiome Project has shed light on how the microbes in and on humans shape our health. A total of 16 projects have been approved, divided among the following categories: Catalog the human virome; Develop tools to study the human virome; Explore how the human virome interacts with us; and, Make virome data available to the research community.
Kyrpides’ project, “Advanced analytics for uncovering virus dynamics and functional potential," is among the five focused on tool development. Specifically, the goal of this work is to develop tools, curated databases, and streamlined workflows that allow researchers to easily analyze viral genomes. This includes (i) classifying viruses into groups that can be compared across studies, (ii) predicting which hosts these viruses are likely to infect, and (iii) identifying the genetic traits of the viruses to better understand their roles and potential impacts. The work is supported by NIH Common Fund grant 1U01DE034196-01.
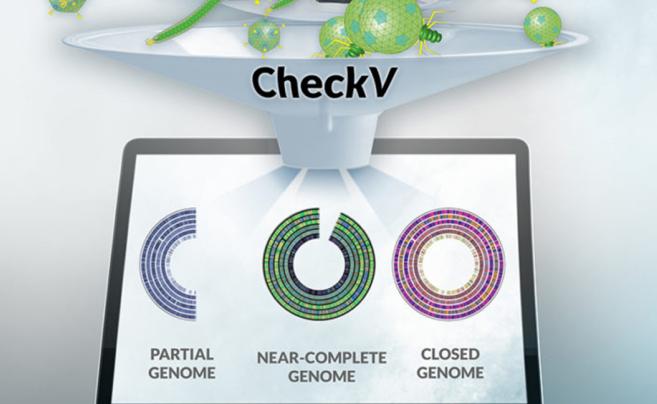
The JGI team, including Kyrpides, Camargo, and Roux, has an extensive track record of developing cutting-edge tools for viral genomics. Notable examples include CheckV, which assesses the quality and completeness of metagenome-assembled viral genomes (Nature Biotechnology, 2021), geNomad, a tool for identifying mobile genetic elements (Nature Biotechnology, 2024), and iPHoP, an integrated machine learning framework to maximize host prediction for metagenome-derived viruses of archaea and bacteria (PLoS Biol, 2023). These tools have significantly advanced the field by providing robust methodologies for analyzing and understanding viral genomes and their interactions with microbial hosts.
The second project, “A consortium facilitation, coordination, and data management center for the HVP," is related to developing an organizational hub for the program, to organize and manage the HVP consortium activities. The hub, named Consortium Organization and Data Collaboration Center (CODCC), is led by a team of highly experienced investigators from institutions including the University of Maryland and Harvard University. The CODCC aims to coordinate data acquisition, analysis, and visualization through a sustainable, state-of-the-art data portal, which will include a database for comparative analysis of all the Human viruses. Kyrpides described this as the development of a human virome specific database of IMG/VR (Viral Resources.) Launched in 2016, IMG/VR is the largest publicly available collection of UViGs by identifying contiguous viral sequences (“contigs”) from thousands of ecologically diverse metagenomics samples. The work is supported by NIH Common Fund grant 1U24HL175772-01.
This is not the first time Kyrpides and his colleagues have demonstrated how tools developed for one field of research can be harnessed elsewhere. In a 2019 paper, the team applied computational methods and analyses developed for JGI-enabled research involving environmental samples to help uncover the phylogenetic diversity of the human gut microbiome.
By bringing their expertise in viral genomics to the Human Virome Program, JGI researchers are similarly poised to contribute transformative insights into the human virome and its implications for health, disease, and biotechnology.