In honor of the JGI's 25th anniversary in 2022, we have revisited a number of notable achievements that showcase our collaborations and capabilities to enable great science that will help solve energy and environmental challenges.
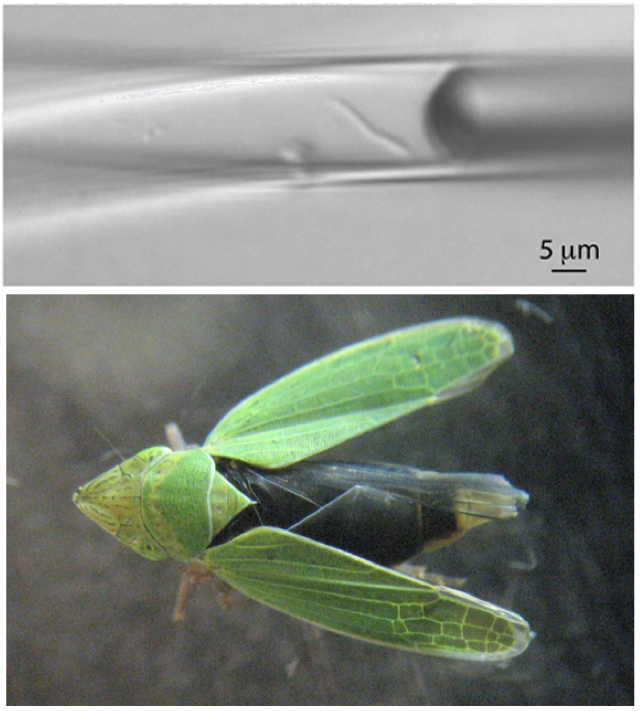
In 2009, researchers at the JGI and the Bigelow Laboratory for Ocean Sciences described a novel technique to access high-quality genomic information using minute quantities of DNA derived from individual environmental cells. The proof of principle study built off earlier approaches and offered a way for researchers to study the planet’s microbial diversity, much of which remains unknown and uncultured in laboratories. Other firsts followed. The next year, the JGI team reported the first closed and finished genome derived from an uncultured bacterial single cell – a symbiont of the green sharpshooter. In 2012, single cells isolated from the microbes found in the oil plume from Deepwater Horizon oil spill led to a draft genome of the first deep-sea, oil-eating bacterium from a single cell. Several years later, the continuing single cell sequencing collaboration between the JGI and Bigelow Lab led to the identification of key microbial culprits in carbon fixation in the dark ocean. Combining single cell genomics and synthetic biology, a team studying microbial populations in oxygen minimum zones identified SAR11 bacteria as initiators of nitrogen loss in these waters.
The top image on the left shows how, from a single bacterial cell (seen here in a glass capillary), researchers sequenced and assembled the first complete genome of an uncultured symbiont. The single cell was isolated from the bacteriome of the green sharpshooter Draeculacephala minerva bacteriome, pictured at the bottom.
Perhaps most notably, JGI’s Microbial Program head Tanja Woyke and her colleagues harnessed the JGI’s single cell genomics capabilities to assemble, identify, and characterize more than 200 microbial genomes that were mapped to nearly 30 major previously uncharted branches of the tree of life. Used in tandem over the years, metagenomics and single cell genomics have afforded JGI users and the larger research community a means to access microbial lineages from a variety of environments. In 2017, the first New Lineages of Life (NeLLi) workshop was held, inviting the research community to discuss how to move from cataloging microbial diversity to experimental characterization and validation of the functional potential of these new lineages.