
SMC is the most comprehensive BGC database available at 10X the scale of the closest comparable database.

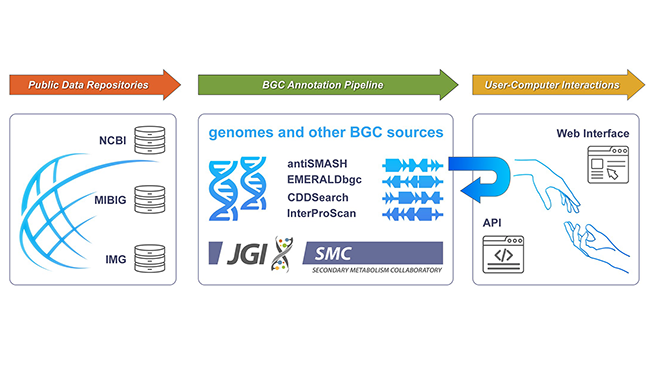
In addition to the core set of "primary metabolites"—chemical compounds shared by most living creatures for energy, growth and reproduction—most organisms produce a set of "secondary metabolites," chemical compounds that often confer specialized capabilities that allow the organisms to adapt to and control their environment. Many secondary metabolites are produced by groups of genes known as biosynthetic gene clusters (BGCs.) As reported in the recent Database issue of the journal Nucleic Acids Research, the Secondary Metabolism Collaboratory (SMC) has launched as an interactive public database for the research community. With more than 13 million BGCs in the catalog at the time of publication, it is the largest repository of BGC sequence data from over 1.3 million publicly available bacterial and archaeal genome and contig sources.
Secondary metabolites are useful in a variety of fields including biotechnology, medicine and agriculture. This comprehensive public repository of BGC sequences that come pre-annotated using multiple commonly used sequence analysis tools is a means of expediting BGC and biosynthesis research.
Over the years several secondary metabolites databases have been made available to the research community, each offering specific features or tools. The U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Berkeley Laboratory, built off past experience in developing a secondary metabolites portal within the Integrated Microbial Genomes and Microbiomes (IMG/M) system known as IMG-ABC. That portal contained biosynthetic gene clusters and secondary metabolites information from isolate and metagenome data.
The SMC (smc.jgi.doe.gov) is designed with two main objectives. First and foremost, it is a BGC repository that aspires to FAIR (findable, accessible, interoperable and reproducible) principles and has tools that allow users to annotate or upload data, as well as to connect and collaborate with each other in a public forum. Second, SMC provides an opportunity for users to publicly engage with each other, share annotations and collaborate on solutions.
Compared with past efforts at the JGI, like IMG-ABC, and other academic efforts, SMC is already the most comprehensive BGC database available at 10X the scale of the closest comparable database, and has the capability to store and accurately display eukaryotic and viral BGC sequences. Expansion of the SMC to include fungal, plant, viral, and metagenome data is currently underway.

