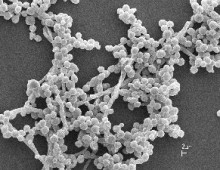
Structural Analysis of Cow and Hoatzin Microbial Communities
Inside the guts of many animals, microbes break down the plant fibers ingested as part of their diet. These microbes are of interest to bioenergy researchers who want to learn from nature and apply these cellulosic degradation capabilities toward biofuel production. To this end, at the JGI, several sequencing projects have focused on the microbial… [Read More]