Signatures of Selection Inscribed on Poplar Genomes
A Nature Genetics study describes a method that could be harnessed for developing more accurate predictive climate change models. [Read More]
 A Nature Genetics study describes a method that could be harnessed for developing more accurate predictive climate change models. [Read More]
A Nature Genetics study describes a method that could be harnessed for developing more accurate predictive climate change models. [Read More] Combining complementary resources for greater scientific understanding. The U.S. Department of Energy Joint Genome Institute (DOE JGI) and the Environmental Molecular Sciences Laboratory (EMSL) have accepted 12 projects submitted during the 2014 call for Collaborative Science Initiative proposals. The collaborative call represents a unique opportunity for researchers to combine the power of genomics and molecular characterization in… [Read More]
Combining complementary resources for greater scientific understanding. The U.S. Department of Energy Joint Genome Institute (DOE JGI) and the Environmental Molecular Sciences Laboratory (EMSL) have accepted 12 projects submitted during the 2014 call for Collaborative Science Initiative proposals. The collaborative call represents a unique opportunity for researchers to combine the power of genomics and molecular characterization in… [Read More]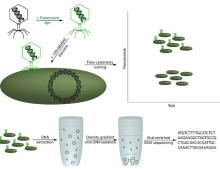 Tagging strategy allows for population surveys The sheer volume of cyanobacteria in the oceans makes them major players in the global carbon cycle and responsible for as much as a third of the carbon fixed. These photosynthetic microbes, which include Prochlorococcus and Synechococcus, are tiny – as many as 100 million cells can be found… [Read More]
Tagging strategy allows for population surveys The sheer volume of cyanobacteria in the oceans makes them major players in the global carbon cycle and responsible for as much as a third of the carbon fixed. These photosynthetic microbes, which include Prochlorococcus and Synechococcus, are tiny – as many as 100 million cells can be found… [Read More] One of the most basic rules for playing the game “Twenty Questions” is that all of the questions must be definitively answered by either “yes” or “no.” The exchange of information allows the players to correctly guess the item in question. Fungal researchers have been using a variation of Twenty Questions to determine if wood-decaying… [Read More]
One of the most basic rules for playing the game “Twenty Questions” is that all of the questions must be definitively answered by either “yes” or “no.” The exchange of information allows the players to correctly guess the item in question. Fungal researchers have been using a variation of Twenty Questions to determine if wood-decaying… [Read More] “Increased to levels unprecedented” is how the Intergovernmental Panel on Climate Change (IPCC) described the rise of carbon dioxide, methane and nitrous oxide emissions in their report on the physical science basis of climate change in 2013. According to the US Environmental Protection Agency (EPA), the atmospheric concentration of methane, a greenhouse gas some 28… [Read More]
“Increased to levels unprecedented” is how the Intergovernmental Panel on Climate Change (IPCC) described the rise of carbon dioxide, methane and nitrous oxide emissions in their report on the physical science basis of climate change in 2013. According to the US Environmental Protection Agency (EPA), the atmospheric concentration of methane, a greenhouse gas some 28… [Read More] From antiseptic oils to the construction of didgeridoos, the traditional Australian Aboriginal wind instrument, the eucalyptus tree serves myriad purposes, accounting for its status as one of the world’s most widely planted hardwood trees. Its prodigious growth habit has caught the eyes of researchers seeking to harness and improve upon Eucalyptus’ potential for enhancing sustainable… [Read More]
From antiseptic oils to the construction of didgeridoos, the traditional Australian Aboriginal wind instrument, the eucalyptus tree serves myriad purposes, accounting for its status as one of the world’s most widely planted hardwood trees. Its prodigious growth habit has caught the eyes of researchers seeking to harness and improve upon Eucalyptus’ potential for enhancing sustainable… [Read More] “It doesn’t take much to see that the problems of three little people doesn’t add up to a hill of beans in this crazy world,” Humphrey Bogart famously said in the movie Casablanca. For the farmers and breeders around the world growing the common bean, however, ensuring that there is an abundant supply of this… [Read More]
“It doesn’t take much to see that the problems of three little people doesn’t add up to a hill of beans in this crazy world,” Humphrey Bogart famously said in the movie Casablanca. For the farmers and breeders around the world growing the common bean, however, ensuring that there is an abundant supply of this… [Read More] Citrus is the world’s most widely cultivated fruit crop. In the U.S. alone, the citrus crop was valued at over $3.1 billion in 2013. Originally domesticated in Southeast Asia thousands of years ago before spreading throughout Asia, Europe, and the Americas via trade, citrus is now under attack from citrus greening, an insidious emerging infectious… [Read More]
Citrus is the world’s most widely cultivated fruit crop. In the U.S. alone, the citrus crop was valued at over $3.1 billion in 2013. Originally domesticated in Southeast Asia thousands of years ago before spreading throughout Asia, Europe, and the Americas via trade, citrus is now under attack from citrus greening, an insidious emerging infectious… [Read More] In the Lewis Carroll classic, Through the Looking Glass, Humpty Dumpty states, “When I use a word, it means just what I choose it to mean—neither more nor less.” In turn, Alice (of Wonderland fame) says, “The question is, whether you can make words mean so many different things.” All organisms on Earth use a genetic code, which is… [Read More]
In the Lewis Carroll classic, Through the Looking Glass, Humpty Dumpty states, “When I use a word, it means just what I choose it to mean—neither more nor less.” In turn, Alice (of Wonderland fame) says, “The question is, whether you can make words mean so many different things.” All organisms on Earth use a genetic code, which is… [Read More] Despite its name, the Dead Sea does support life, and not just in the sense of helping visitors float in its waters. Algae, bacteria, and fungi make up the limited number of species that can tolerate the extremely salty environment at the lowest point on Earth. Some organisms thrive in salty environments by lying dormant… [Read More]
Despite its name, the Dead Sea does support life, and not just in the sense of helping visitors float in its waters. Algae, bacteria, and fungi make up the limited number of species that can tolerate the extremely salty environment at the lowest point on Earth. Some organisms thrive in salty environments by lying dormant… [Read More]